Page last modified: Monday, 24-Oct-2022 10:22:09 EDT
 |
 |
The dataset for tables beyond 111 markers is reduced from the page 1 dataset as not all Y111 testers have completed a Big Y test.
Clicking a cluster label should pop up project notes for that cluster. The cluster labels are tan rows starting with a number. If clicking the row does not work, click the NOTES icon in the toolbar.
The Compare utility is enabled for this haplogroup. Using the checkboxes at the very far left of the haplogroup table, you can select any members in the table and juxtapose them for comparison in a separate window.
Table #3 Haplotypes for I-P37 Driscolls Table #3 Haplotypes for I-P37 Driscolls | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 203 | 204 | 205 | 206 | 207 | 208 | 209 | 210 | 211 | 212 | 213 | 214 | 215 | 216 | 217 | 218 | 219 | 220 | 221 | 222 | 223 | 224 | 225 | 226 | 227 | 228 | 229 | 230 | 231 | 232 | 233 | 234 | 235 | 236 | 237 | 238 | 239 | 240 | 241 | 242 | 243 | 244 | 245 | 246 | 247 | 248 | 249 | 250 | 251 | 252 | 253 | 254 | 255 | 256 | 257 | 258 | 259 | 260 | 261 | 262 | 263 | 264 | 265 | 266 | 267 | 268 | 269 | 270 | 271 | 272 | 273 | 274 | 275 | 276 | 277 | 278 | 279 | 280 | 281 | 282 | 283 | 284 | 285 | 286 | 287 | 288 | 289 | 290 | 291 | 292 | 293 | relative position | |||||||||||
| Short Tandem Repeat Markers (STRS) | DYF398B | FTY43 | FTY166 | FTY13 | FTY11 | DYS584 | DYS608 | FTY95 | FTY151 | FTY388 | FTY256 | DYS580 | FTY234 | FTY268 | FTY139 | FTY27 | FTY75 | DYS512 | FTY329 | FTY320 | FTY7 | FTY247 | FTY211 | DYS474 | FTY39 | FTY376 | FTY37 | FTY373 | DYS475 | FTY138 | FTY288 | FTY264 | FTY93 | FTY380 | FTY18 | FTY115 | DYS569 | FTY390 | FTY46 | FTY243 | FTY322 | FTY281 | FTY153 | FTY181 | FTY184 | FTY359 | FTY121 | FTY74 | FTY279 | DYS530 | FTY45 | DYS573 | DYS542 | FTY36 | FTY304 | FTY203 | FTY291 | FTY142 | FTY191 | FTY183 | FTY141 | FTY299 | FTY193 | FTY124 | FTY16 | FTY236 | FTY185 | FTY378 | FTY225 | FTY397 | FTY3 | FTY67 | FTY182 | FTY334 | DYS623 | FTY348 | FTY357 | FTY275 | FTY306 | FTY383 | FTY253 | FTY285 | DYS645 | FTY17 | FTY85 | DYS598 | FTY375 | FTY325 | FTY12 | FTY365 | FTY35 | Short Tandem Repeat Markers (STRS) | |||||||||||
| member # |
|
member # | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
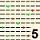
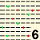
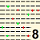
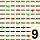
| MODAL | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | MODAL | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17102828 I-Y64698 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 10 | - | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | - | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | - | 8 | 6 | 11 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 11 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 17102828 I-Y64698 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 70494771 I-A13665 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 11 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 12 | 7 | 5 | 9 | 5 | 5 | 4 | 11 | 8 | 6 | 10 | 9 | - | 4 | 4 | 5 | 5 | 70494771 I-A13665 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 97290794 I-BY133999 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 12 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 11 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 11 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 97290794 I-BY133999 | |||||||||||
| 20580628 I-BY133999 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 11 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | - | 5 | 9 | 5 | 5 | 4 | 11 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 20580628 I-BY133999 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 29219807 I-FT46638 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 11 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 29219807 I-FT46638 | |||||||||||
| 40842301 I-FT46638 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | - | 8 | 5 | - | 6 | 4 | 4 | 4 | 5 | - | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | - | 6 | 4 | 9 | 4 | - | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 13 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 40842301 I-FT46638 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 28974321 I-Y37101 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 28974321 I-Y37101 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81166406 I-Y112683 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | - | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 81166406 I-Y112683 | |||||||||||
| 53000774 I-Y112683 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | - | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | - | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | - | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | - | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | - | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 53000774 I-Y112683 | |||||||||||
| 62832597 I-Y112683 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | - | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | - | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 62832597 I-Y112683 | |||||||||||
| 80447821 I-A14359 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 80447821 I-A14359 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 68239442 I-BY37208 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | - | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 68239442 I-BY37208 | |||||||||||
| 74603646 I-BY123243 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 74603646 I-BY123243 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 53096021 I-A19162 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 12 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 53096021 I-A19162 | |||||||||||
| 69620219 I-A19162 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 69620219 I-A19162 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 75851244 I-BY90072 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | - | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 13 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 75851244 I-BY90072 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10499705 I-Y100290 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 13 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | - | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 10499705 I-Y100290 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 75851244 I-BY90072 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | - | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 13 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 75851244 I-BY90072 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 90532743 I-BY54260 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 13 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 90532743 I-BY54260 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 84644678 I-BY54260 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 11 | 4 | 5 | 5 | 8 | 6 | 14 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 13 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 84644678 I-BY54260 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16690339 I-BY86789 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | - | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 16690339 I-BY86789 | |||||||||||
| 21072300 I-BY86789 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 21072300 I-BY86789 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 59530551 I-BY176167 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 13 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 11 | 9 | 5 | 4 | 4 | 5 | 5 | 59530551 I-BY176167 | |||||||||||
| 56746798 I-BY176167 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 56746798 I-BY176167 | |||||||||||
| 51579115 I-BY176167 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | - | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 51579115 I-BY176167 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13095018 I-BY176167 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 12 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | - | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 13095018 I-BY176167 | |||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 18995386 I-FT3387 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | - | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 11 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 18995386 I-FT3387 | |||||||||||
| 15117514 I-FT3387 | 5 | 5 | 5 | 5 | 5 | 8 | 8 | 6 | 5 | 6 | 6 | 9 | 5 | 4 | 4 | 5 | 5 | 8 | 5 | 5 | 5 | 4 | 5 | 8 | 4 | 4 | 4 | 4 | 8 | 5 | 5 | 6 | 4 | 4 | 4 | 5 | 11 | 4 | 6 | 6 | 6 | 4 | 4 | 4 | 4 | 5 | 6 | 6 | 4 | 9 | 4 | 10 | 12 | 4 | 5 | 5 | 8 | 6 | 11 | 6 | 5 | 11 | 5 | 6 | 6 | 5 | 8 | 4 | 4 | 4 | 6 | 4 | 4 | 5 | 11 | 7 | 5 | 9 | 5 | 5 | 4 | 12 | 8 | 6 | 10 | 9 | 5 | 4 | 4 | 5 | 5 | 15117514 I-FT3387 | |||||||||||
| member # |
| member # | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 203 | 204 | 205 | 206 | 207 | 208 | 209 | 210 | 211 | 212 | 213 | 214 | 215 | 216 | 217 | 218 | 219 | 220 | 221 | 222 | 223 | 224 | 225 | 226 | 227 | 228 | 229 | 230 | 231 | 232 | 233 | 234 | 235 | 236 | 237 | 238 | 239 | 240 | 241 | 242 | 243 | 244 | 245 | 246 | 247 | 248 | 249 | 250 | 251 | 252 | 253 | 254 | 255 | 256 | 257 | 258 | 259 | 260 | 261 | 262 | 263 | 264 | 265 | 266 | 267 | 268 | 269 | 270 | 271 | 272 | 273 | 274 | 275 | 276 | 277 | 278 | 279 | 280 | 281 | 282 | 283 | 284 | 285 | 286 | 287 | 288 | 289 | 290 | 291 | 292 | 293 | relative position | |||||||||||
| Short Tandem Repeat Markers (STRS) | DYF398B | FTY43 | FTY166 | FTY13 | FTY11 | DYS584 | DYS608 | FTY95 | FTY151 | FTY388 | FTY256 | DYS580 | FTY234 | FTY268 | FTY139 | FTY27 | FTY75 | DYS512 | FTY329 | FTY320 | FTY7 | FTY247 | FTY211 | DYS474 | FTY39 | FTY376 | FTY37 | FTY373 | DYS475 | FTY138 | FTY288 | FTY264 | FTY93 | FTY380 | FTY18 | FTY115 | DYS569 | FTY390 | FTY46 | FTY243 | FTY322 | FTY281 | FTY153 | FTY181 | FTY184 | FTY359 | FTY121 | FTY74 | FTY279 | DYS530 | FTY45 | DYS573 | DYS542 | FTY36 | FTY304 | FTY203 | FTY291 | FTY142 | FTY191 | FTY183 | FTY141 | FTY299 | FTY193 | FTY124 | FTY16 | FTY236 | FTY185 | FTY378 | FTY225 | FTY397 | FTY3 | FTY67 | FTY182 | FTY334 | DYS623 | FTY348 | FTY357 | FTY275 | FTY306 | FTY383 | FTY253 | FTY285 | DYS645 | FTY17 | FTY85 | DYS598 | FTY375 | FTY325 | FTY12 | FTY365 | FTY35 | Short Tandem Repeat Markers (STRS) | |||||||||||