 |
 |
 SNP Calendar / Timeline
SNP Calendar / Timeline
- R1b1a1b1a1a2c1a3a2 CTS4466 200 C.E.
- R1b1a1b1a1a2c1a3a2a S1115 200 C.E.
- R1b1a1b1a1a2c1a3a2a1 A541 200 C.E.
- R1b1a1b1a1a2c1a3a2a1a S1121 200 C.E.
- R1b1a1b1a1a2c1a3a2a1a1 L270/Z16520 850 C.E. remote SULLIVAN
- R1b1a1b1a1a2c1a3a2a1a1? BY29108 2nd mill
- R1b1a1b1a1a2c1a3a2a1a2 Y6238/Z16252 450 C.E.
- R1b1a1b1a1a2c1a3a2a1a2. Y18975/A9005 1000 C.E.
- R1b1a1b1a1a2c1a3a2a1a2.. A6516 2nd mill
- R1b1a1b1a1a2c1a3a2a1a2... BY99310 unknown L CHRONICANE
- R1b1a1b1a1a2c1a3a2a1a2.. A6516 2nd mill
- R1b1a1b1a1a2c1a3a2a1a2. Y18975/A9005 1000 C.E.
- R1b1a1b1a1a2c1a3a2a1a1 L270/Z16520 850 C.E. remote SULLIVAN
- R1b1a1b1a1a2c1a3a2a1b A195 600 C.E.
- R1b1a1b1a1a2c1a3a2a1b1 A761
- R1b1a1b1a1a2c1a3a2a1b1a A88 700 C.E.
- R1b1a1b1a1a2c1a3a2a1b1a1 Z16259 700 C.E.
- R1b1a1b1a1a2c1a3a2a1b1a1. A10727 L
- R1b1a1b1a1a2c1a3a2a1b1a1c A2220
- R1b1a1b1a1a2c1a3a2a1b1a1c. A7755 L
- R1b1a1b1a1a2c1a3a2a1b1a1 Z16259 700 C.E.
- R1b1a1b1a1a2c1a3a2a1b1a A88 700 C.E.
- R1b1a1b1a1a2c1a3a2a1b1 A761
- R1b1a1b1a1a2c1a3a2a1a S1121 200 C.E.
- R1b1a1b1a1a2c1a3a2a1 A541 200 C.E.
- R1b1a1b1a1a2c1a3a2a S1115 200 C.E.
The Driscolls here fall into R1b subclade R-CTS4466 and also meet the broader definition of the subclade formerly known as South Irish now used in the R-L21 South Irish Project. Newer research hypothesizes that descendant branches of R-CTS4466 moved directly from the Iberian peninsula to Scotland and Northern Ireland.
R-CTS4466 has a major presence in Cork. The last time the Cork Ireland Regional DNA Project charted the haplogroups of its male project members claiming a Cork direct paternal line, R-CTS4466 constituted some 38% of all the men who were chartable. Roughly 20% of Driscoll project members fall or are thought to fall within R-CTS4466.
In 2019, we have identified a third R-CTS4466 branch among our Driscolls, downstream of R-Z16259.
If you are willing to do sufficient STR and SNP testing, please join the R1b-CTS4466 Plus Haplogroup Project.
Several R-CTS4466 Driscolls have been inactive in the project for well over a year. They may bear yet more information but we need more fully active participants. There is too little information on R-CTS4466 Driscolls to draw many conclusions!
R-CTS4466 members are in private project Clusters 4500 - 4565.
 Haplotypes Page 1 Back to top
Haplotypes Page 1 Back to top
The modal haplotype in the table below is not calculated from the data of active members. It is the Irish Type II modal haplotype borrowed from the Munster Irish project. The project member records are then compared to the broader modal.
The Driscolls in the different subclades of R-CTS4466 most likely obtained their surname in a different way rather than from a single R-CTS4466 man named Driscoll. So a good Irish Type II modal haplotype is the best modal haplotype choice when comparing records.
However, no published Irish Type II modal haplotype out to 561 markers or 838 markers is known to exist. Therefore, the additional haplotype pages will show an extended modal calculated from the active members who have done advanced Big Y testing.
Clicking a cluster label should pop up project notes for that cluster. The cluster labels are tan rows starting with a number. If clicking the row does not work, click the NOTES icon in the toolbar.
| Table #1 Haplotypes for R-CTS4466 Driscolls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 1 | 2 | 3 | 4 | 5-6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14-15 | 16 | 17 | 18 | 19 | 20 | 21 | 22-25 | 26 | 27 | 28-29 | 30 | 31 | 32 | 33 | 34-35 | 36 | 37 | 38 | 39 | 40-41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49-50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | relative position | |||||||||
| Short Tandem Repeat Markers (STRS) | DYS393 | DYS390 | DYS19 | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | Short Tandem Repeat Markers (STRS) | |||||||||
| member # |
|
member # | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MODAL | 13 | 24 | 14 | 10 | 11-15 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-17 | 11 | 11 | 19-23 | 15 | 15 | 18 | 17 | 36-37 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 21 | 15 | 19 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | MODAL | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 66957373 R-BY29108 | 13 | 24 | 14 | 10 | 11-16 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 25 | 15 | 19 | 29 | 15-15-17-17 | 11 | 11 | 19-23 | 15 | 15 | 19 | 17 | 36-38 | 12 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-25 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 23 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 34 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 12 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 19 | 14 | 24 | 17 | 12 | 15 | 25 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 66957373 R-BY29108 | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12710740 R-BY99310 | 13 | 24 | 14 | 10 | 11-16 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10-10 | 11 | 11 | 24 | 14 | 19 | 29 | 15-15-16-16-17-17 | 11 | 11 | 19-21 | 16 | 15 | 18 | 17 | 36-36-36 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 15 | 10 | 12 | 12 | 15 | 8 | 12 | 23 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 34 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 15 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 21 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 12710740 R-BY99310 | |||||||||
| 31198509 R-M269 | 13 | 24 | 14 | 10 | 11-16 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10-10 | 11 | 11 | 24 | 14 | 19 | 29 | 15-15-16-16-17-17 | 11 | 11 | 19-21 | 16 | 15 | 18 | 17 | 36-36-36 | 14 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 15 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 33 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 15 | 12 | 11 | 9 | 12 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 21 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 31198509 R-M269 | |||||||||
| 47034272 R-M269 | 13 | 24 | 14 | 10 | 11-16 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 14 | 19 | 29 | 15-15-16-17 | 11 | 11 | 19-21 | 16 | 15 | 18 | 17 | 36-36 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 15 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 33 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 15 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 21 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 47034272 R-M269 | |||||||||
| 47173224 R-M269 | 13 | 24 | 14 | 11 | 11-16 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 14 | 19 | 29 | 15-15-16-17 | 11 | 12 | 19-21 | 16 | 15 | 18 | 17 | 36-36 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 15 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 33 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 15 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 21 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 47173224 R-M269 | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 54332621 R-A10727 | 13 | 24 | 14 | 9 | 11-15 | 12 | 12 | 11 | 13 | 13 | 29 | 16 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 14-15-17-17 | 11 | 11 | 19-23 | 16 | 15 | 18 | 16 | 36-38 | 12 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 16 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 15 | 12 | 23 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 12 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 21 | 15 | 19 | 14 | 25 | 16 | 12 | 15 | 24 | 12 | 24 | 19 | 10 | 14 | 18 | 8 | 12 | 11 | 54332621 R-A10727 | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 70045411 R-M269 | 13 | 24 | 14 | 10 | 10-16 | 12 | 12 | 12 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-17 | 11 | 12 | 19-23 | 16 | 15 | 18 | 17 | 36-40 | 12 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 12 | 12 | 11 | 9 | 12 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 24 | 15 | 19 | 14 | 25 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 70045411 R-M269 | |||||||||
| 39499302 R-A7755 | 13 | 24 | 14 | 10 | 11-16 | 12 | 12 | 12 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-17 | 11 | 12 | 19-23 | 16 | 14 | 19 | 17 | 36-39 | 12 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 12 | 12 | 11 | 9 | 12 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 25 | 15 | 19 | 14 | 25 | 15 | 13 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | 39499302 R-A7755 | |||||||||
| member # |
| member # | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
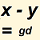 Genetic Distance Back to top
Genetic Distance Back to top
|
Since the modal is now a broader R-CTS4466 modal haplotype rather than a project-calculated data, individual members will overall show a slightly greater distance from that modal. The first tester after MODAL is in the Beara Driscoll/Sullivan group. Cluster 4550 has Driscoll matches who are not actively participating. Cluster 4555 members are Chronicane Driscolls (Sherkin Island and Ardfield). We see extra copies on more than one multi-copy marker in some Chronicane data. The software used to calculate Genetic Distance now counts the occurrence of extra copies on multi-copy markers DYS459, DYS464, and CDY as one mutation. The rationale for this is in Part III of the tutorial, discussed under palindromic multi-copy markers. Hopefully this yields a more realistic Genetic Distance. Refer also to "Genetic Distance" in the haplogroup help guide. |
|
Genetic Distance
Color Key
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
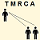 TMRCA Table Back to top
TMRCA Table Back to top
| TMRCA for R-CTS4466 Driscolls Experimental | ||||||||||||||
| ID | MODAL | 66957373 R-BY29108 | 12710740 R-BY99310 | 31198509 R-M269 | 47034272 R-M269 | 47173224 R-M269 | 54332621 R-A10727 | 70045411 R-M269 | 39499302 R-A7755 | |||||
| MODAL | 111 | 14-35 | 17-40 | 20-45 | 17-40 | 20-45 | 18-42 | 15-37 | 21-47 | |||||
| 66957373 R-BY29108 | 14-35 | 111 | 21-47 | 25-52 | 25-52 | 28-57 | 28-57 | 23-49 | 26-54 | |||||
| 12710740 R-BY99310 | 17-40 | 21-47 | 111 | 1-11 | 2-13 | 4-18 | 37-70 | 30-59 | 37-70 | |||||
| 31198509 R-M269 | 20-45 | 25-52 | 1-11 | 111 | 2-13 | 4-18 | 37-70 | 30-59 | 37-70 | |||||
| 47034272 R-M269 | 17-40 | 25-52 | 2-13 | 2-13 | 111 | 1-11 | 37-70 | 30-59 | 37-70 | |||||
| 47173224 R-M269 | 20-45 | 28-57 | 4-18 | 4-18 | 1-11 | 111 | 40-75 | 30-59 | 37-70 | |||||
| 54332621 R-A10727 | 18-42 | 28-57 | 37-70 | 37-70 | 37-70 | 40-75 | 111 | 25-52 | 31-62 | |||||
| 70045411 R-M269 | 15-37 | 23-49 | 30-59 | 30-59 | 30-59 | 30-59 | 25-52 | 111 | 7-23 | |||||
| 39499302 R-A7755 | 21-47 | 26-54 | 37-70 | 37-70 | 37-70 | 37-70 | 31-62 | 7-23 | 111 | |||||
| ||||||||||||||
 Phylogenetic Tree / Analysis Diagrams Back to top
Phylogenetic Tree / Analysis Diagrams Back to top
|
See the Archives page for older diagrams. The diagram below shows a vertical calendar on the right edge, starting from 200 C.E. This is a timeline of SNP formation dates. (The calendar at the top of this page shows TMRCA estimates.) If you need a refresher on the difference between the two, refer to the tutorial, part 3, at or about question #27. Several SNP streams might say mutations not yet identified or mutation date unknown. These remind us that there are tremendous gaps in what we know. Mutations not yet identified means the project expects considerably further SNP definition along the stream. Mutation date unknown is shown with a dashed line. It means we don't how to pin an SNP formation date precisely to the timeline bar on the right side of the diagram. 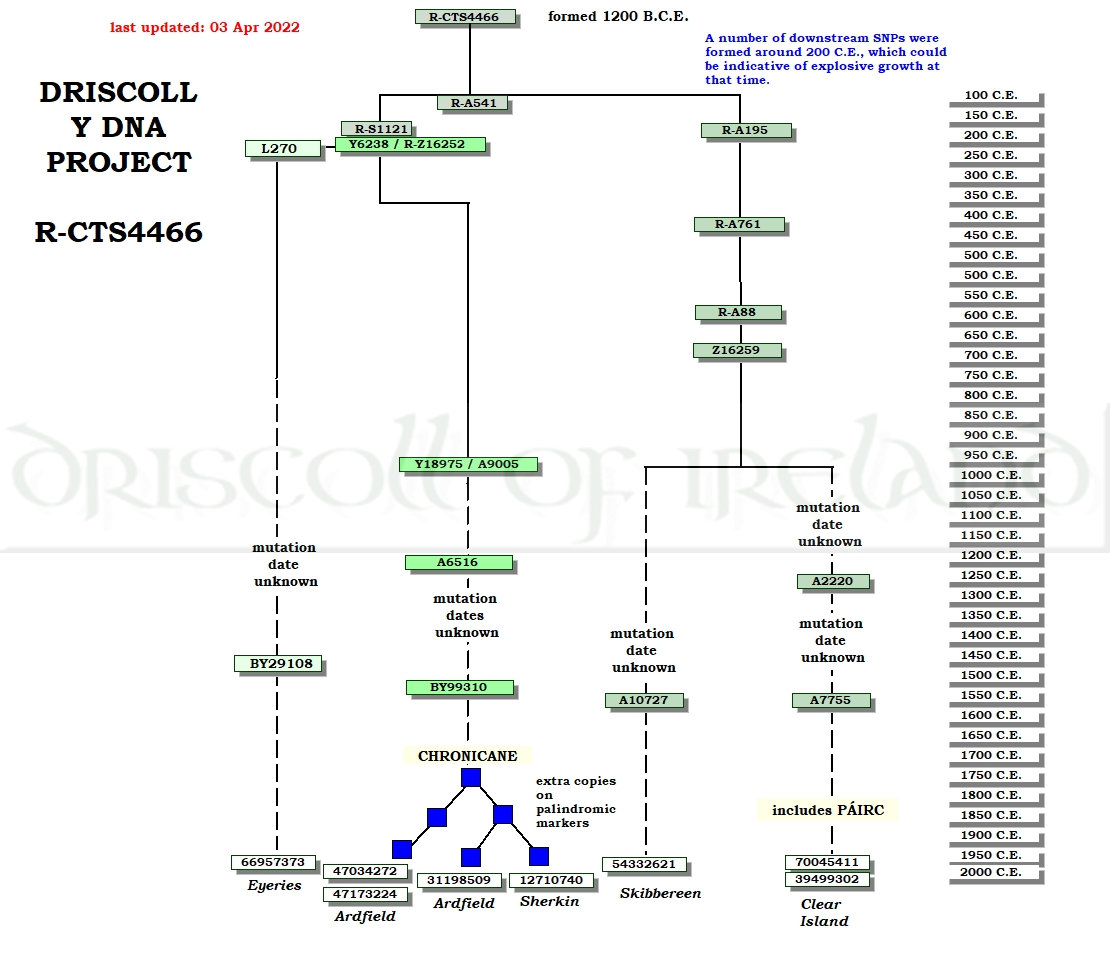 |